ALS Unraveled: A patient-centric approach to drug discovery and clinical validation
Executive Summary
Amyotrophic Lateral Sclerosis (ALS) remains one of medicine’s most challenging neurodegenerative diseases, with an over 95% clinical trial failure rate leading to only modestly effective therapies that extend life by months, not years. Time remains one of the most devastating barriers in ALS research: drug development takes 10-15 years on average while most diagnosed patients live only 2-5 years. Since many biological insights primarily come from post-mortem tissue, current ALS patients are unlikely to receive timely, actionable therapeutic benefits for their condition. This is not a failure of effort, funding, or expertise; it reflects a systemic challenge in how ALS therapeutics are currently developed.
Unravel offers a new approach. By using real-time RNA profiles from accessible samples (nasal swabs from current patients) to build a “living molecular twin” of inaccessible neurological tissues, Unravel models each ALS patient’s unique, dynamic, system-wide problem. This same digital patient avatar allows not only for the prediction of customized therapeutic mechanisms, but these new mechanisms can then be rapidly tested using available approved drugs. This is Unravel’s Predictable Medicine™, which enables discovery to start with each patient and end with clinical validation in the very same patient in a matter of 2-3 months. The goal is to bridge foundational mechanistic research to patients: turning biological knowledge into timely, actionable treatment paths for patients who participate in ALS research. Unravel aims to bring real options to people living with ALS now, shifting ALS drug discovery to delivering solutions in the present.
Background
Despite over a century of dedicated research, amyotrophic lateral sclerosis (ALS) remains one of the most devastating and least tractable neurological diseases. The condition was first described in 1869 by Jean-Martin Charcot [1], yet no curative therapy exists today. Once symptoms appear, median survival is only 2 to 5 years, and fewer than 10% of patients live beyond a decade [2]. Each year, around 5,000 new cases are diagnosed in the United States alone, and the global prevalence continues to rise with aging populations and improved diagnostics [3].
From a therapeutic standpoint, progress has been painfully slow. Across nearly one thousand clinical trials, over 95% of ALS drug candidates have failed [4], and only four drugs: Riluzole (1995), Nuedexta (2010), Edaravone (2017), and Tofersen (2023) have achieved U.S. FDA approval [5]. Notably, they offer limited benefit, typically extending survival by just 3 to 6 months [3], and are not compatible treatment options for all those diagnosed with ALS. For instance, Tofersen is only suitable for around 2% of the diagnosed population [5]. Moreover, estimates of median survival from diagnosis remain in the range of 2 to 3 years, underscoring the urgency for novel therapeutic options for patients alive today. [6]
Despite decades of global investment, ALS research has seen limited therapeutic progress relative to the scale of funding. Since the 2014 Ice Bucket Challenge alone, [7] the ALS Association has committed over $160 million to more than 560 projects worldwide [8], while U.S. federal funding has grown to roughly $143 million annually through the NIH [9], and nearly $200 million through broader congressional allocations in 2022 [10]. Altogether, this represents billions of dollars invested over the past two decades by public, private, and philanthropic sources yet only a handful of modestly effective drugs have emerged, with average survival after diagnosis remaining at two to five years [3]. The challenge is not insufficient effort or resources, but rather the lack of dynamic, systems-level data capable of decoding ALS biology in patients and rapidly testing on those very same patients in a timeframe that can benefit them directly.
Unravel’s Predictable Medicine™ framework offers a new path forward: by using dynamic RNA-based network modeling, we can move beyond static, single-gene models of disease and instead map how ALS disrupts the entire biological system across tissues, time, and disease stages.
This whitepaper outlines how Unravel’s BioNAV™ systems biology platform can:
Stratify patients into specific treatable subgroups even if the cause is unknown.
Outline therapeutic prediction for ALS by reframing the disease as a biological network imbalance, not an isolated neuronal failure.
Accelerate drug development to get to the clinic faster.
First, understanding the core challenges in ALS research and why progress has lagged is crucial to identifying a solution. Cancer, a similarly devastating disease, has seen transformative advances over the past 20 years by embracing molecular profiling, patient stratification, and biomarker-driven precision medicine. Therapeutic development for ALS, however, has remained constrained by technical barriers that make real-time, patient-specific biology difficult to access and interpret. These limitations have slowed translation from biological insight to clinical impact. Understanding these gaps highlights where new approaches like Unravel’s systems-biology model can finally move the sector forward from a field rich in hypotheses to one rich in clinically validated solutions.
ALS Needs
Biological heterogeneity
Resulting Limitations
ALS is not one disease; it manifests differently across the patient population yet is treated as one.
Poor patient stratification
Except for a few genetically defined subgroups, clinical trials group patients by symptoms, not molecular state, leading to inconsistent results and failed trials.
Most biomarkers currently in use are static, late-stage, or derived from postmortem tissues which make them not reflective of true disease activity in living patients.
Limited biomarkers
High failure in clinical trials
Clinical trials are money- and time-intensive, and over 95% of clinical trials for current drugs have failed. There are only 4 approved drugs for ALS after 20+ years of global research.
Unravel addresses these challenges with its network-based approach [11-13]. By analyzing RNA signatures from non-invasively collected samples like nasal swabs, Unravel’s AI platform builds living molecular twins of patients. These living molecular twins are used to rapidly screen thousands of molecules that restore systems-level balance, uncover entirely new biology with the potential to transform ALS treatment, and validate the discoveries using readily-available drugs in the very same patients – all in 2-3 months. Target-centric research on SOD1, C9ORF72, NEK [14], and others remains essential for understanding mechanisms but is inherently time-consuming, sometimes focused only on a subset of patients, and historically hard to translate into the clinic. To translate insight into benefit for patients sooner, mechanistic discovery must be paired with clinical validation methods like Unravel’s that prioritize speed, actionability, and efficacy in living patients. In practice, Unravel’s “living molecular twins” operate symbiotically with academic research: our patient-level predictions guide what to test first, creating real-time clinical validation opportunities for patients in the present, while mechanistic studies explain why and how those interventions work and can be improved in the future.
Given the following problem: ALS research studies fragments of a system, one piece at a time, stopping and examining every piece to understand where it fits in the puzzle.
Unravel presents this solution: Unravel models the system itself, focusing on high level function and working towards getting the system back to equilibrium.
Empirical Support: Unravel’s Dynamic, Network-Based, Action-Oriented Approach
RNA Used as a Dynamic Readout
RNA reflects the real-time biological state - how the body is responding to the stress of the disease, the environment (i.e., diet, activity, sleep conditions, air quality), and any treatments currently in use.
Cross-Tissue Network Modelling
Using our BioNAV™ AI platform, Unravel reconstructs in silico how genes, proteins, and pathways interact across tissues. A neuronal dysfunction alters immune or metabolic genes which can be captured through the nasal swabs that Unravel uses to collect transcriptomic data. These accessible samples become faithful surrogates for the neuronal and system-level damage ALS has caused within the patient.
Together, the following analyses demonstrate how BioNAV™ network modeling extracts consistent, cross-tissue disease and therapeutic mechanisms and reveals patterns that are not apparent from raw gene expression alone.
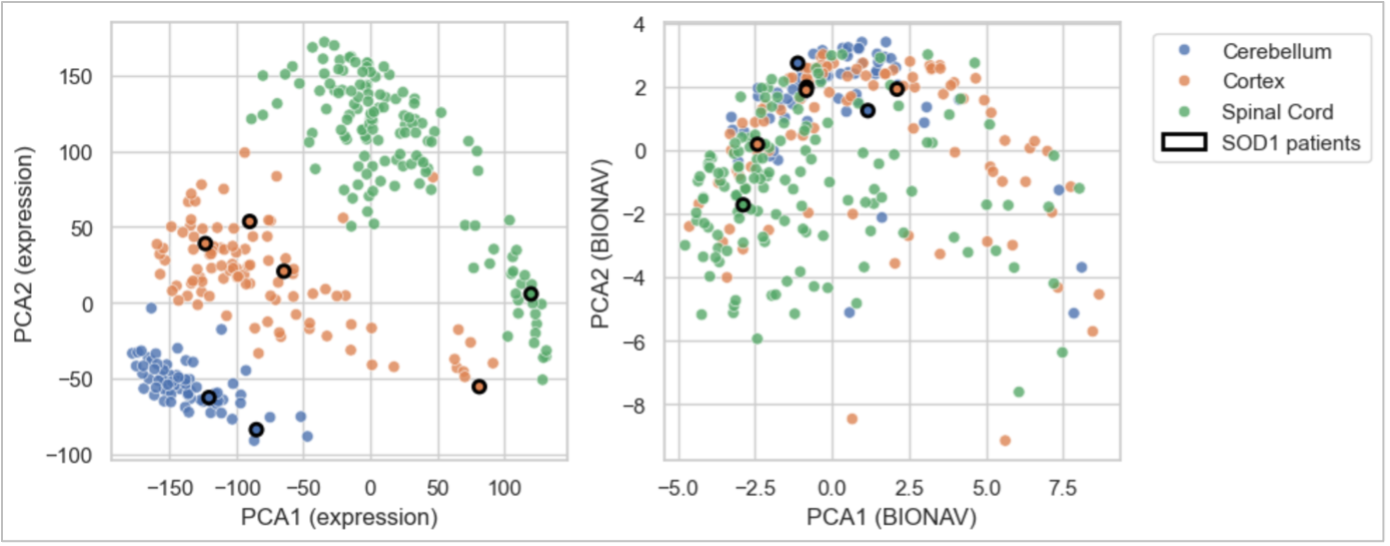
Figure 1. Cross-Tissue Transcriptome Translation Better Highlights Molecular Mechanisms of Disease
The left figure shows the principal component analysis of raw gene expression across samples from cortex, spinal cord, and cerebellum, where even molecularly similar SOD1 patients remain separated by tissue. In contrast, the BioNAV™ network-based platform (right) transforms these tissue-driven expression signatures into therapeutic pathway-driven representations, where SOD1 samples are now closer to each other. This demonstrates that despite tissue-specific differences in raw expression, the underlying therapeutic response signals are consistent for similar patients, despite the tissue. Importantly, BioNAV™ can detect and extract those cross-tissue molecular mechanisms relevant to ALS biology.
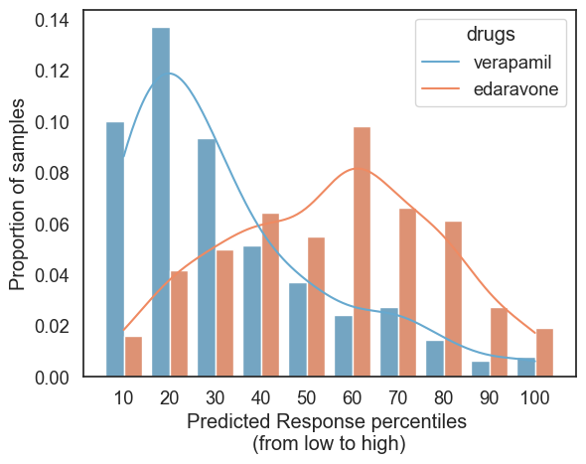
Figure 2. BioNAV™ recapitulates literature-supported therapeutic differences while capturing patient heterogeneity
BioNAV™ recapitulates known efficacy differences between ALS therapies while revealing substantial patient-level heterogeneity. For each RNA profile, BioNAV™ predicts whether that patient would benefit from a drug (Rank 100%) or would have an adverse effect (0%). This figure shows that across the ALS population, most patients show low predicted benefit for verapamil, which is consistent with its failure in clinical studies. In contrast, the approved drug edaravone displays a broader distribution of predicted responses, with a subset of patients showing higher predicted benefit. This stratification highlights how BioNAV™ organizes heterogeneous patient biology into therapeutically relevant subgroups, enabling the detection of population-level trends while preserving individual variation. These patterns align with literature-reported differences in these drugs’ performances and illustrate BioNAV™’s ability to identify who is more or less likely to benefit, rather than assuming uniform response across all patients with a shared diagnosis. Together, these results demonstrate BioNAV™’s power to generate predictable, mechanism-driven therapeutic insights at both the subgroup and individual-patient level.
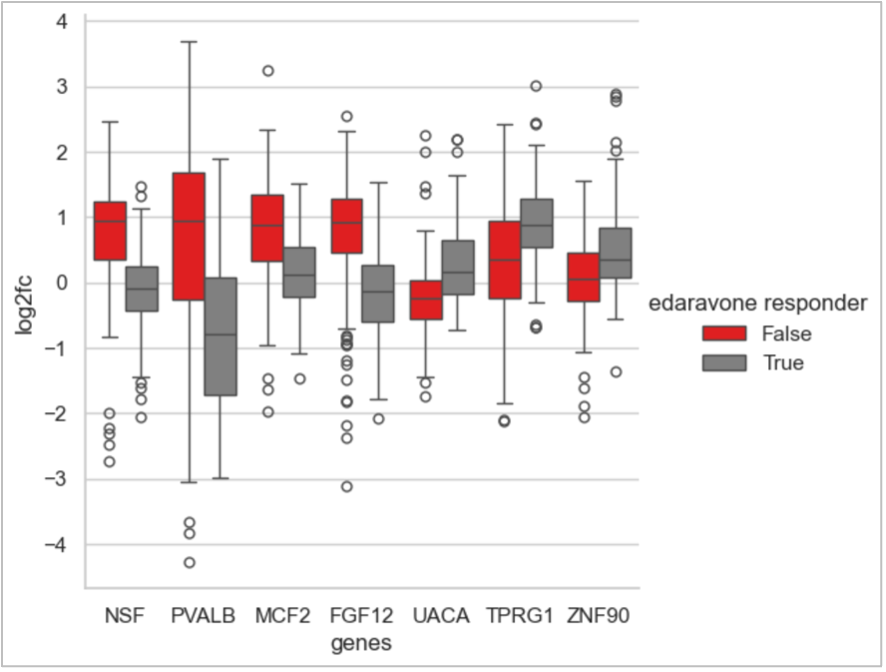
Figure 3. BioNAV™ Identifies Novel Cross-Tissue, Neurodegeneration-Linked Indicators of Drug Response
This figure shows a curated set of gene-level indicators that differentiate predicted edaravone non-responders (red) from responders (grey). After BioNAV™ stratifies patients into response groups, analysis identifies the genes most associated with these response differences. Many of these indicators are linked in the literature to neurodegenerative biology and brain-related pathways, and importantly, they emerge consistently across tissues rather than being tissue-specific. This demonstrates how BioNAV™ reveals the mechanistic drivers underlying predicted drug response, enabling clear responder vs. non-responder stratification and highlighting the system-level signals that connect central and peripheral biology.
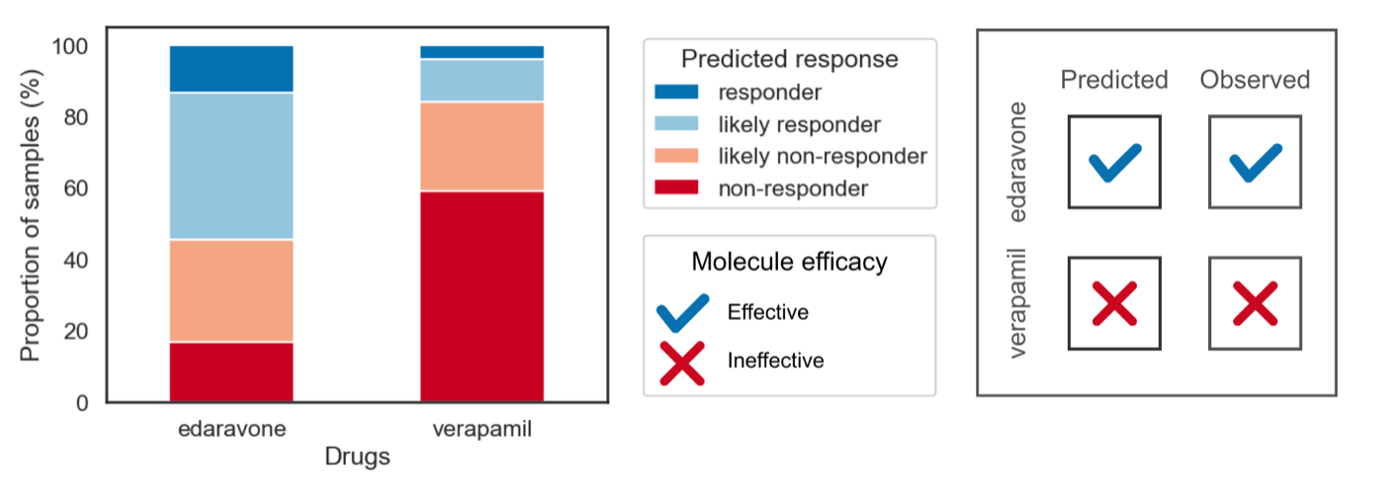
Figure 4. BioNAV™ Correctly Predicts Outcomes of ALS Clinical Trials Outcomes
These figures demonstrate BioNAV™’s ability to correctly predict the efficacy of existing ALS candidate treatments. BioNAV™ predicts over 80% of ALS patients will be likely non-responders or likely non-responders to verapamil. Similarly, the observed clinical trial outcomes of verapamil were that it is an ineffective treatment for ALS [15]. BioNAV™ also predicts over 50% of the ALS population will either be responders or likely responders to edaravone. This aligns with clinical trial outcomes, where edaravone was observed to be beneficial to ALS patients [16]. BioNAV™ achieved similar estimates using RNA profiling alone. This demonstrates that BioNAV™ can be used to predict the clinical efficacy of future ALS candidate treatments.
Together, these analyses illustrate how BioNAV™ overcomes core challenges in ALS by capturing biological heterogeneity, stratifying patients into mechanistically meaningful subgroups, and identifying novel cross-tissue indicators of drug response. By resolving the molecular complexity that has historically obscured ALS drug development, BioNAV™ enables biological network-targeted drug candidates to enter better-designed, more efficient clinical trials that increase the likelihood of therapeutic success.
Discussion
Unravel addresses key ALS drug discovery and development needs through the “living molecular twin” in the following ways:
Each patient is analyzed as their own molecular system. By comparing individual RNA profiles against Unravel’s database of healthy controls, the BioNAV™ platform identifies which pathways are disrupted in that specific patient, enabling personalized treatment predictions instead of a one-size-fits-all disease model. Importantly, this is not a “one patient, one drug” personalized medicine that fails to scale. By identifying recurring network signatures across many patients, Unravel can group individuals into biologically defined subtypes: creating targeted, scalable treatment strategies while still respecting ALS heterogeneity.
ALS Needs
Biological heterogeneity
Unravel’s Solution
Poor patient stratification
Using network clustering, Unravel groups patients by molecular mechanism rather than symptom onset. This identifies mechanistic subtypes (e.g., inflammation-dominant vs. mitochondrial-dominant ALS), improving trial design and matching patients to the most promising therapies.
Limited biomarkers
RNA-based network signatures from non-invasive samples (nasal swabs) capture real-time disease activity. These dynamic biomarkers can be tracked over time to measure disease progression or therapeutic response: addressing the lack of living, longitudinal markers in ALS.
High failure in clinical trials
The BioNAV™ platform enables in silico testing of hundreds of repurposed compounds before proof-of-concept human trials with available drugs, allowing rapid elimination of ineffective options. Unravel accelerates candidate selection and reduces clinical trial risk, time, and cost.
Conclusion
ALS has long been viewed through the wrong lens: as a neurological mystery instead of a systemic imbalance. Unravel’s approach makes it possible to see how the disease unfolds not just in neurons but across the body. By connecting transcriptomic data with network-level insights, Unravel aims to bring Predictable Medicine™ to the ALS community: accelerating discovery, stratifying trial design, and ultimately, effective treatments.
Authorship
This paper was authored by Aruzhan Bekbolatova, Mikayla Reitsma, Viola Fanfani, and Richard Novak.
References
1. History of ALS | The ALS Association. Accessed October 28, 2025. https://www.als.org/understanding-als/lou-gehrig.
2. About amyotrophic lateral sclerosis (ALS). Centers for Disease Control and Prevention. Accessed October 28, 2025. https://www.cdc.gov/als/abouttheregistrymain/about-amyotrophic-lateral-sclerosis-als.html#:~:text=Although%20no%20one%20knows%20for,doctors%20to%20report%20ALS%20cases.
3. Masrori P, Van Damme P. Amyotrophic lateral sclerosis: A clinical review. European Journal of Neurology. 2020;27(10):1918-1929. doi:10.1111/ene.14393
4. Kuldip D. 1 update on ALS therapeutic landscape and ALS Association’s ... Update on ALS Therapeutic Landscape and ALS Association’s strategy to have more, better and faster trials. Accessed October 28, 2025. https://www.als.org/sites/default/files/2025-02/ALS Therapeutic Landscape and ALS Association’s Strategy to Have More, Better, and Faster Trials.pdf.
5. Medications for treating ALS | the ALS Association. Navigating ALS. Accessed October 28, 2025. https://www.als.org/navigating-als/living-with-als/medications.
6. Engelberg-Cook E, Shah JS, Teixeira da Silva Hucke A, et al. Prognostic factors and epidemiology of amyotrophic lateral sclerosis in southeastern United States. Mayo Clinic Proceedings: Innovations, Quality & Outcomes. 2024;8(5):482-492. doi:10.1016/j.mayocpiqo.2024.07.008
7. Chapman M. Ice bucket challenge greatly aided ALS research, study reports. ALS News Today. June 10, 2019. Accessed October 28, 2025. https://alsnewstoday.com/news/ice-bucket-challenge-dramatically-affected-als-fight/.
8. Abnowf R, Bernacet A, Fletcher J, et al. New report highlights progress made because of als ice bucket challenge | The ALS Association. Navigating ALS. Accessed October 28, 2025. https://www.als.org/stories-news/new-report-highlights-progress-made-because-als-ice-bucket-challenge.
9. Federal public policy priorities | The ALS Association. Federal Public Policy Priorities. Accessed October 28, 2025. https://www.als.org/advocacy/our-priorities/federal-public-policy-priorities.
10. Federal budget bill boosts spending on ALS Research | The ALS Association. Federal Budget Bill Boosts Spending on ALS Research. Accessed October 28, 2025. https://www.als.org/blog/federal-budget-bill-boosts-spending-als-research.
11. Flores R, Nihalani R, Umur S, Vigneault F, Novak R. Drug-gene network signature modeling predicts breast cancer patient response to neoadjuvant chemotherapy. Published online March 18, 2025. doi:10.21203/rs.3.rs-6130021/v1
12. Novak R, Lin T, Kaushal S, et al. Ai-enabled drug prediction and gene network analysis reveal therapeutic use of vorinostat for rett syndrome in preclinical models. Communications Medicine. 2025;5(1). doi:10.1038/s43856-025-00975-8
13. Sperry MM, Oskotsky TT, Marić I, et al. Target-agnostic drug prediction integrated with medical record analysis uncovers differential associations of statins with increased survival in COVID-19 patients. PLOS Computational Biology. 2023;19(5). doi:10.1371/journal.pcbi.1011050
14. Nijs M, Van Damme P. The genetics of amyotrophic lateral sclerosis. Current Opinion in Neurology. 2024;37(5):560-569. doi:10.1097/wco.0000000000001294
15. Miller RG;Smith SA;Murphy JR;Brinkmann JR;Graves J;Mendoza M;Sands ML;Ringel SP; A clinical trial of verapamil in amyotrophic lateral sclerosis. Muscle & nerve. Accessed November 12, 2025. https://pubmed.ncbi.nlm.nih.gov/8622731/.
16. Shefner J, Heiman‐Patterson T, Pioro EP, et al. Long‐term edaravone efficacy in amyotrophic lateral sclerosis: Post‐Hoc Analyses of study 19 (MCI186‐19). Muscle & Nerve. 2019;61(2):218-221. doi:10.1002/mus.26740
©Unravel Biosciences 2025 | Contact@unravel.bio
| BioNAV™, RARIFY™, Predictable Medicine™ are exclusive trademarks of Unravel Biosciences |